 |
| Figure 1B from Baym's science paper (link above). The real size is on the order of feet... that is a LOT of bugs. |
What it would mean is that instantly, all 10^6 or so bugs would change their genotype... well, this was clearly crazy wrong :)
So, we started thinking about how populations spread in space (Fisher-like waves), and we found the wonderful body of work from the Gore, Hallatschek and Korolev labs thinking about mutational surfing (essentially at the wave tip there is a changed effective population size so the balance between drift and selection can change)
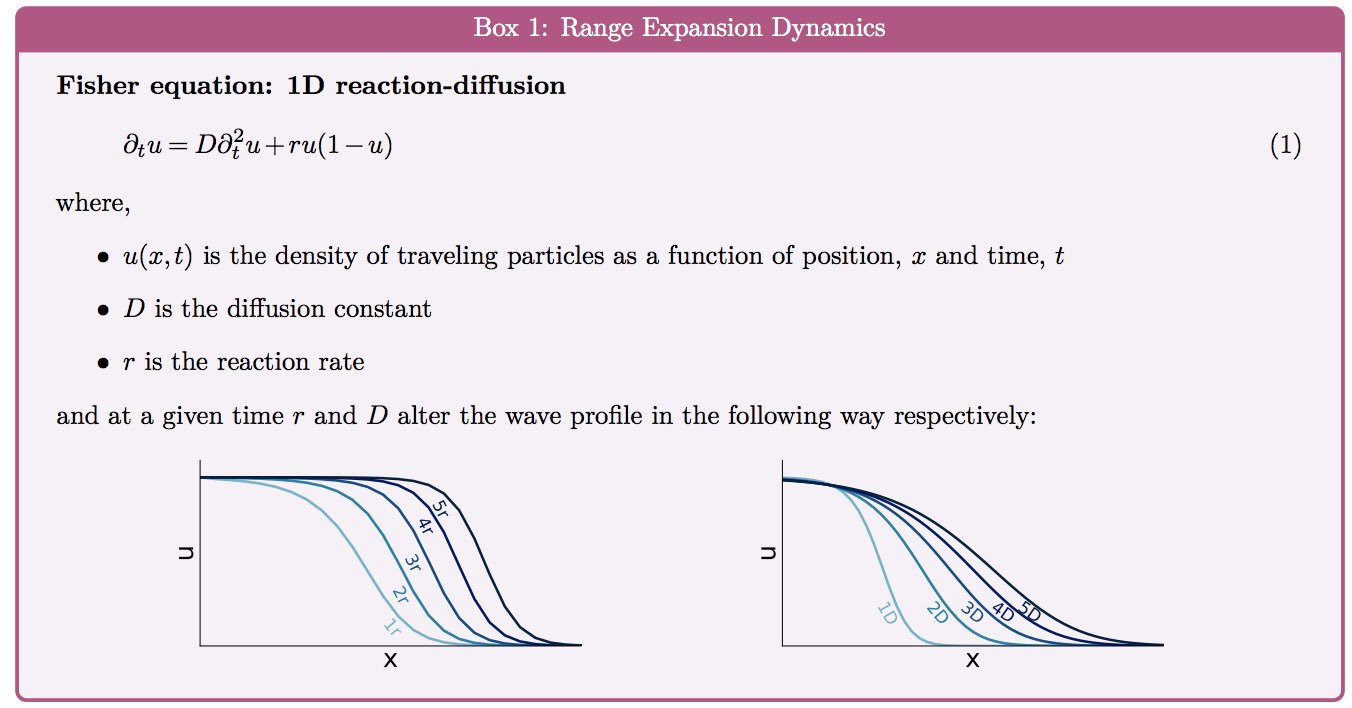
Formulating a model of (stochastic) mutation dynamics in a Fisher-like wave like those before us yielded known results. For a range of parameters, fixation of mutants can be promoted near the wave front - often called "mutations surfing on a wave".
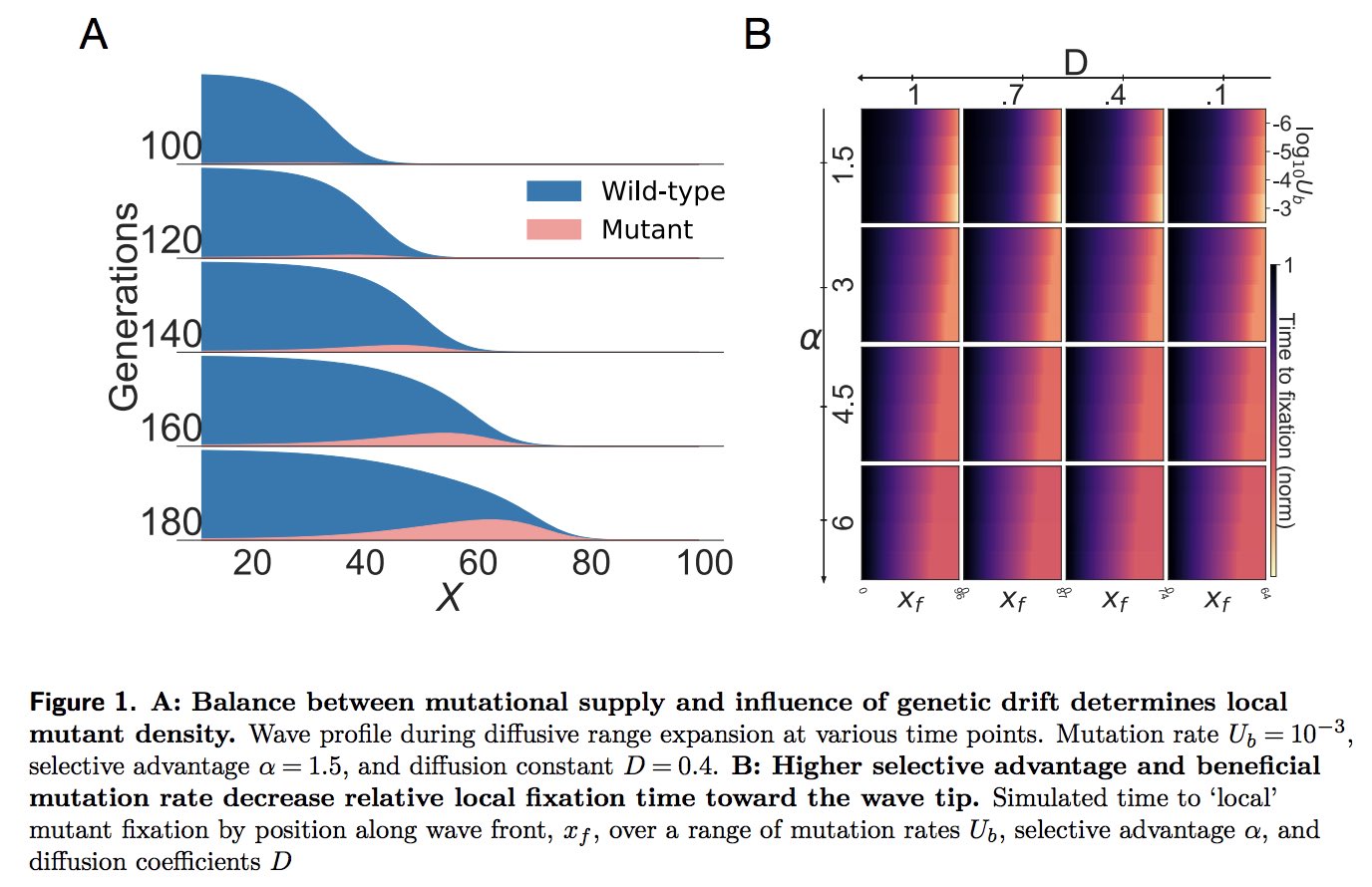
So far, all this is known. Let's also recall from standard (well mixed) popgen (think Kimura) that depending on the relationship between the average time to establishment of a mutant, and the time to fixation, we can have qualitatively different evolutionary dynamics (e.g. strong selection weak mutation (SSWM) when the time to establishment is far larger than time to fixation; and weak selection strong mutation (WSSM) or strong selection strong mutation (SSSM) when the opposite is true, of if they are comparable). This is where we started to wonder...
we think a lot about strong selection (like in the form of experimental evolution in antibiotics or associated mathematical models of similar systems) and often make the assumption that our dynamics are in the SSWM regime -- but this need not always be the case (in fact, it likely isn't).
So, to really get at this, Nikhil defined a 'clonal interference index' as the log of the ratio of the fixation and establishment time, and calculated this all along the wavefront. What popped out is a pretty fun result: depending where are you on the wave front, you experience qualitatively different evolutionary dynamics.
This is our first real effort working in the range expansion space -- so we are very eager for any feedback (in particular if we've missed key references or if this is already a solved problem!).
Please check out the full preprint on bioRxiv for more details, and drop us a line if you have comments/questions.




